
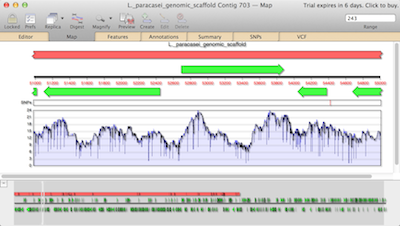
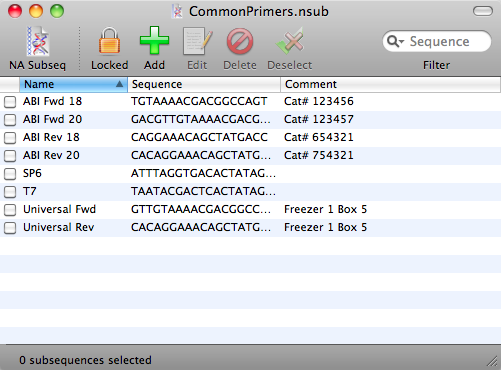
The output options include lists of sites found and sites not found, the subsequence map, the subsequence annotated sequence, and the fragment predictions. The results dialog box for nucleic acid subsequence analysis is similar to that of restriction analysis. The analysis itself is done in a fashion similar to restriction analysis, by selecting the AnalyzeiNucleic Acid Subsequence command. The new subsequence file can be saved and then used in nucleic acid subsequence analysis. Select subsequences can be highlighted by Shift-clicking, copied to the clipboard and pasted into a new nuc1eic acid subsequence file (opened by the FileiNew command and selecting Nucleic subsequence from the pull-down menu). As in the restriction enzyme files, each row of a subsequence file consistS of the name, the recognition site, and any additional information (such as references) of a nucleic acid subsequence. each of which can be opened using the FileiOpen command. MacVecror comes with six nucleic acid subsequence files. Fragments can be predicted using single or double digests. The map can be toggled between circular and linear by clicking on the circular/linear button in the Graphics Palette. The restriction map can be customized to include features and the results can be filtered so that only a subset of the cut sites is shown. Once the analysis is complete, the results dialog box lists the output options: a list of restriction enzymes that cut, a list of those that do not, the restriction map, the cut site annotated sequence, and finally the fragment prediction. The pull-down menu next to Search using offers the user the choice of using all enzymes in the restriction enzyme file or only the enzymes that were selected earlier. The user clicks on the Enzyme File button to select a restriction enzyme file for the analysis. Selecting the AnalyzeiRestriction Enzyme command brings up the restriction analysis dialog box. Clicking on an enzyme puts a check mark at the left of its name, and selects this enzyme for use in restriction analysis. The FileiOpen command is used to open the restriction enzyme files: Each row has the name, cut site, and any known isoschizomers of a restriction enzyme (Fig. The restriction enzyme files are lists of enzymes carried by different manufacturers. which consists of several restriction enzyme files. Restriction and Motif Analyses MacVector comes with the REBASE restriction enzyme database. The Filter button in the Graphics Palette for the Results Map can also be used to filter the results.ģ.5. or line for Hnear maps or radius of circle for circular maps), line length for linear displays. The Graphics Palette also allows control of the display density of residues (number of residues per inch, centimeter. For DNA sequences, the user can toggle between linear and circular display by selecting the linear/circular button. 3) allows the user to switch the individual map elements on or off, e.g., the user may decide that a certain feature should not be shown on the map or that only a particular restriction site should be shown. the list includes Title, RuJer, Sequence, and Features, whereas for a Results Map the list includes these items and Results. The scrolling list in this dialog box lists the individual map elements whose appearance can be edited. the Graphics Palette dialog box is also displayed. The check boxes adjacent to the individual map elements enable the user to switch the elementsidut>st( inchġ81 ウセャゥ、エᆳ OsrHinU' l8l r・ウオャエセ@ 181 ATilt 1818dmllt ャォエセ@ĭow.
The graphics palette for the circular and linear forms of the p♻R322 restriction map. Adrmtr&K Receptor Protoc:ols, edittd by Otr1is A. Mokrlllar M.dhocls 111 Developrntllt-' Bioiocr: Xeoopus and Ztbrofish. lntetrill Protoroh, ・、ゥエセ@ by Anthony Howle//, 1999 f28. Ttmlaiplion Ftttor Prol«ok, ediled by セゥャイエョ@ J. llioinfonutlcs Methods ond Ptococols, edited by Sup/len MlsefiK ntH! Stephen A.
Gene Tl'ltlift& Protocols, edited by £ric B. T Cdl Prot.ocols: Dew/opmlfrl and Ac:tivotiOtt, edited by Ktlly P. Pl111t Hormone Protocols, edlltd by Jert11'1 A. Transformin& Growth Ftdor-Belll Protocols. Protein Strutlure l'rtdiclion: Mtlho4 11nd Protorols. Ctlpain Metlloclsand Prototols.tdittd by JohnS. Baeterial ToliM: Malw4s 1111d Pl'tJtocols: ediltd by Olto Holst. M E T H 0 D S I N M 0 l E C U l A R B I 0 L 0 G Y"'


 0 kommentar(er)
0 kommentar(er)
